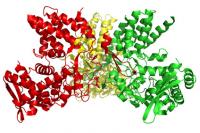
The atomic structure of influenza A virus nucleoprotein. Credit: Jane Tao/Rice University Striking new research from Rice University and the University of Texas at Austin (UT) has revealed a potential new target that drug makers can use to attack several strains of influenza, including those that cause bird flu as well as the common variety that infects millions each flu season.
The research, published online today by Nature, offers tantalizing evidence of a potential drug target in a flu protein called nucleoprotein, or NP. NP plays a vital role in all strains of influenza A, including Hong Kong flu, Spanish flu and bird flu. __IMAGE_2
The target is NP's long, flexible tail. Biochemists at Rice and UT found that even minor changes to the tail prevented NP from fulfilling one of its roles – linking together into structural columns that the virus uses to transmit copies of itself.
"There is a small binding pocket for the tail loop of the protein that appears to be a promising target for a new class of antiviral drugs," said lead researcher Jane Tao, assistant professor in biochemistry and cell biology. "We know from previous genetic studies that this tail loop is almost identical across strains of influenza A, so drugs that target the tail have a high potential of being effective against multiple strains, including the H5N1 strains. Such new antivirals are especially needed at the moment as some H5N1 viruses are resistant to the flu drug Tamiflu."
Tao's findings are based on a painstaking series for experiments that revealed the atomic structure of NP. The protein's structure was discerned via X-ray crystallography, a method that allows scientists to discern the three-dimensional placement of atoms in a crystal based upon the diffraction patterns of X-rays that pass through it.
Tao said it was a challenge to growing NP protein crystals. The method used was the hanging drop vapor diffusion method, which involves suspending a liquid droplet of concentrated protein solution on the underside of a glass slide that is sealed inside a jar. As the liquid in the droplets evaporates, the proteins become supersaturated, and in some cases they form tiny crystals of a few hundred microns in size. Tao estimates that postdoctoral research associate Qiaozhen Ye prepared about 1,000 jars, with multiple droplets per jar, to get the 100 or so crystals that were needed for the experiments.
NP is one of only 11 proteins that are encoded by the influenza A genome. One of its main functions is structural. Once the virus has hijacked a host cell, and converted it into a virus-replicating factory, the NPs come together in small rings as building blocks. Many NP rings stack one atop the other in a slightly off-registered fashion, forming long helical-shaped columns. The virus's RNA genome is twisted around this column and shipped out to infect other cells.
"NP has about 500 amino acids and the tail loop contains about 30 of those," Tao said. "We found that a mutation in only one residue out of 30 was enough to prevent the NPs from coming together to form the building blocks for the columns, and without these columns the virus cannot make copies and infect other cells."
Tao said the research also provides clues about NP's role in signaling a cell to begin making copies of the viral genome, and Tao's group is continuing its work with co-author Robert Krug at the University of Texas at Austin to explore the protein's regulatory functions. Source : Rice University
 Print Article
Print Article Mail to a Friend
Mail to a Friend
