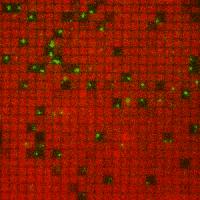
This gif of membrane-anchored Ras (red) and individual SOS molecules (green) shows individual SOS molecules corralled into nanofabricated patches where all the membrane-associated Ras molecules they activate can be trapped. A breakthrough discovery into how living cells process and respond to chemical information could help advance the development of treatments for a large number of cancers and other cellular disorders that have been resistant to therapy. An international collaboration of researchers, led by scientists with the U.S. Department of Energy (DOE)'s Lawrence Berkeley National Laboratory (Berkeley Lab) and the University of California (UC) Berkeley, have unlocked the secret behind the activation of the Ras family of proteins, one of the most important components of cellular signaling networks in biology and major drivers of cancers that are among the most difficult to treat.
"Ras is a family of membrane-anchored proteins whose activation is a critical step in cellular signaling, but almost everything we know about how Ras signals are activated has been derived from bulk assays, in solution or in live cells, in which information about the role of the membrane environment and anything about variation among individual molecules is lost," says Jay Groves, a chemist with Berkeley Lab's Physical Biosciences Division and UC Berkeley's Chemistry Department. "Using a supported-membrane array platform, we were able to perform single molecule studies of Ras activation in a membrane environment and discover a surprising new mechanism though which Ras signaling is activated by Son of Sevenless (SOS) proteins."
Groves, who is also a Howard Hughes Medical Institute (HHMI) investigator, is the corresponding author of a paper in Science that reports this discovery. The paper is titled "Ras activation by SOS: Allosteric regulation by altered fluctuation dynamics." The lead authors were Lars Iversen and Hsiung-Lin Tu, both members of Groves' research group at the time of the study. See below for a complete list of co-authors and their institutional affiliations.
The cellular signaling networks of living cells start with receptor proteins residing on a cell's surface that detect and interact with the environment. Signals from these receptors are transmitted to chemical networks within the cell that process the incoming information, make decisions, and direct subsequent cellular activities. __IMAGE_2
"Although cellular signaling networks perform logical operations like a computer microprocessor, they do not operate in the same way," Groves says. "The individual computational steps in a standard computer are deterministic; the outcome is determined by the inputs. For the chemical reactions that compose a cellular signaling network, however, the molecular level outcomes are defined by probabilities only. This means that the same input can lead to different outcomes."
For cellular signaling networks involving large numbers of protein molecules, the outcome can be directly determined by the process of averaging. Even though the behavior of an individual protein is intrinsically variable, the average behavior from a large group of identical proteins is precisely determined by molecular level probabilities. Ras activation in a living cell, however, involves a relatively small number of SOS molecules, making it impossible to average the variable behavior of the individual molecules. This variation is referred to as stochastic "noise" and has been widely viewed by scientists as an error a cell must overcome.
"Our study showed that, in fact, an important aspect of the SOS signal that activates Ras is encoded in the noise," says Groves. "The protein's dynamic fluctuations between different states of activity transmit information, which means we have found a regulatory coupling in a protein signaling reaction that is entirely based on dynamics, without any trace of the signal being seen in the average behavior."
The Ras Enigma
Ras proteins are essential components of signaling networks that control cellular proliferation, differentiation and survival. Mutations in Ras genes were the first specific genetic alterations linked to human cancers and it is now estimated that nearly a third of all human cancers can be traced to something going wrong with Ras activation. Defective Ras signaling has also been cited as a contributing factor to other diseases, including diabetes and immunological and inflammatory disorders. Despite this long history of recognized association with cancers and other diseases, Ras proteins have been dubbed "un-druggable," largely because their activation mechanism has been poorly understood.
A roadblock to understanding Ras signaling is that the membranes to which Ras proteins are anchored play a major role in their activation through SOS exchange factors. SOS activity in turn was believed to be allosterically regulated through protein and membrane interactions, but this was deduced from cell biological studies rather than direct observations. For a better understanding of how Ras activation by SOS is regulated, scientists need to observe individual SOS molecules interacting with Ras in a membrane environment. However, membrane environments have traditionally presented a stiff experimental challenge. __IMAGE_3
Groves and his research group overcame this challenge with the development of supported membrane arrays constructed out of lipid layers embedded with fixed patterns of metal nanostructures and assembled onto a silica substrate. The metal structures allow for the controlled spacing of proteins and other cellular molecules placed on the membranes. This makes it possible for the membranes to serve as a platform for assays that can be used to observe in real-time the activity of single molecules.
"In this case, our supported membrane allowed us to corral individual SOS molecules into nanofabricated patches that trapped all the membrane-associated Ras molecules they activated," Groves says. "This in turn allowed us to monitor the individual contribution of every molecule in the ensemble and reveal how the dynamic transitions of individual molecules encoded information that is lost in the average."
What the collaboration discovered is that SOS regulation is based on the dynamics of distinct stochastic fluctuations between different activity states that last approximately 100 seconds but do not show up in ensemble averages. These long-lived fluctuations provide the mechanism of allosteric SOS regulation and Ras activation.
"The allosteric regulation of SOS deduced from cell biological and bulk biochemical studies is conspicuously absent in direct single molecule studies," Groves says. "This means that something that was inferred to exist proved to be missing when we did an experiment that explicitly measured it. The dynamic fluctuations we observed within the system correlated with the expected allosteric regulation, and subsequent theoretical modeling confirmed that such stochastic fluctuations can give rise to known bulk effects."
Understanding the role of stochastic dynamic fluctuations as signaling transduction mechanisms for Ras proteins, could point the way to new and effective therapies for Ras-driven cancers and other cellular disorders. In their Science paper, the collaborators also express their belief that the dynamic fluctuations mechanism they discovered is not unique to Ras proteins but could be applicable to a broad range of other cellular signaling proteins.
"The reason this mechanism has not been reported before is that no previous experiment could have revealed it," Groves says. "All previous experiments on this system - and most others for that matter - were based on average behavior. Only single molecule measurements that can look at all the molecules in the system are capable of revealing this type of effect, which we think may prove to be very important in the function of living cell signaling systems."
Source : lcyarris@lbl.gov
 Print Article
Print Article Mail to a Friend
Mail to a Friend
