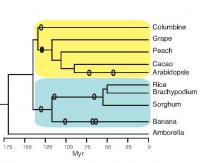
This image shows the evolutionary relationships among the species analyzed for conserved non-coding sequences. 'Myr' stands for million years ago. Ellipses are approximate times of whole-genome duplications. DNA is the molecule that encodes the genetic instructions enabling a cell to produce the thousands of proteins it typically needs. The linear sequence of the A, T, C, and G bases in what is called coding DNA determines the particular protein that a short segment of DNA, known as a gene, will encode. But in many organisms, there is much more DNA in a cell than is needed to code for all the necessary proteins. This non-coding DNA was often referred to as "junk" DNA because it seemed unnecessary. But in retrospect, we did not yet understand the function of these seemingly unnecessary DNA sequences.
We now know that non-coding DNA can have important functions other than encoding proteins. Many non-coding sequences produce RNA molecules that regulate gene expression by turning them on and off. Others contain enhancer or inhibitory elements. Recent work by the international ENCODE (Encyclopedia of DNA Elements) Project (1, 2) suggested that a large percentage of non-coding DNA, which makes up an estimated 95% of the human genome, has a function in gene regulation. Thus, it is premature to say that "junk" DNA does not have a function—we just need to find out what it is!
To help understand the importance of this large amount of non-coding DNA in plants, Diane Burgess and Michael Freeling at the University of California, Berkeley have identified numerous conserved non-coding sequences (CNSs) of DNA that are found in a wide variety of plant species, including rice, banana, and cacao. DNA sequences that are highly conserved, meaning that they are identical or nearly so in a variety of organisms, are likely to have important functions in basic biological processes. For example, the gene encoding ribosomal RNA, an essential part of the protein-synthesizing machinery needed by cells of all organisms, is highly conserved. Changes in the sequence of this key molecule are poorly tolerated, so ribosomal RNA sequences have changed relatively little over millions of years of evolution.
To identify the most highly conserved plant CNSs, Burgess and Freeling compared the genome (one copy of all the DNA in an organism) of the model plant Arabidopsis, a member of the mustard family, with the genome of columbine, a distantly related plant of the buttercup family. The phylogenetic tree (see figure) shows the evolutionary relationships among the dicot (yellow) and monocot (blue) species they studied. Branch points represent points of divergence of two species from a common ancestor. Sequences in common between these two plants, which diverged over 130 million years ago, are likely to have important functions or they would have been lost due to random mutations or insertions or deletions.
They found over 200 CNSs in common between these distantly related species. In addition, 59 of these CNSs were also found in monocots, which are even more distant evolutionarily, and these were termed deep CNSs. Finally, they showed that 51 of these appear to be found in all flowering plants, based on their occurrence in Amborella, a flowering plant that diverged from all of the above plants even before the monocot-dicot split (see figure).
So what could be the function of these deep CNSs? We can get clues by analyzing the types of genes with which these CNSs are associated. The researchers found that nearly all of the deep CNSs are associated with genes involved in basic and universal biological processes in flowering plants—processes such as development, response to hormones, and regulation of gene expression. They found that the majority of these CNSs are associated with genes involved in tissue and organ development, post-embryonic differentiation, flowering, and production of reproductive structures. Others are associated with hormone- and salt-responsive genes or with genes encoding transcription factors, which are regulatory proteins that control gene expression by turning other genes on and off.
In addition, they showed that these CNSs are enriched for binding sites for transcription factors, and propose that the function of some of this non-coding DNA is to act as a scaffold for organization of the gene expression machinery. The binding sites they found are known sequences implicated in other plants as necessary for response to biotic and abiotic stress, light, and hormones. Furthermore, they discovered that a number of the CNSs could produce RNAs that have extensive double-stranded regions. These double-stranded regions have been shown to be involved in RNA stability, degradation, and in regulation of gene expression. Twelve of the most 59 highly conserved CNSs are associated with genes whose protein products interact with RNA. Clearly, these DNA sequences are not merely "junk!"
Now that Burgess and Freeling have identified the most highly conserved non-coding DNA sequences in flowering plants, future scientists have a better idea of which regions of the genome to focus on for functional studies. Do the predicted transcription factor-binding sites actually bind known or novel transcription factors? Do CNSs organize or regulate the gene expression machinery? Do CNSs encode RNAs that regulate fundamental processes in plants? The answers to these and many related questions will be easier to answer now that we have this set of deep CNSs that are likely to play important roles in basic cellular processes in plants. Source : tspady@aspb.org
 Print Article
Print Article Mail to a Friend
Mail to a Friend
