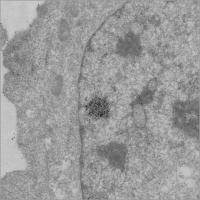
The new technique exposes living cells to labeled antibodies, an approach that yields a much stronger signal for electron microscopy. University of Illinois researchers have developed a technique for imaging cells under an electron microscope that yields a sharper image of the structure of chromatin, the tightly wound bundle of genetic material and proteins that makes up the chromosomes.
Their findings appear in Nature Methods.
Scientists have known for more than a century that proteins, such as histones, aid in packing DNA into the nucleus of a cell. Human cells contain 2 to 3 meters of DNA, which must be kinked and coiled enough to fit into a region 1/10 the width of a human hair.
Despite the use of powerful, high-resolution imaging techniques such as electron microscopy, the mechanism by which this chromatin packing occurs remains a mystery. The densely coiled chromatin fibers are very difficult to visualize, and little is known about how they condense during cell division, or unwind to allow gene expression.
In developing their method, the Illinois team tackled a key difficulty in imaging cells using electron microscopy. Traditional studies “fix” the cells with potent chemicals (called fixatives) to preserve their structure for viewing under a microscope. But standard fixation methods interfere with another step in the imaging process: the use of tagged antibodies to label key components of the cells. 
The new technique exposes living cells to labeled antibodies, an approach that yields a much stronger signal for electron microscopy.
These antibodies, which target and latch on to specific proteins in the cell, can be tagged with fluorescent labels for detection in light microscopy, or with metal particles (gold, in this case) for electron microscopy.
“If you fix the cells first, you have a dramatic drop in the efficiency of these immunochemical reactions,” said Igor Kireev, a visiting scientist in the department of cell and developmental biology and lead author of the paper.
“And if your target is inside the condensed chromatin, the antibodies have no way to penetrate.”
Instead of fixing the cells before staining with antibodies, the researchers first exposed living animal cells to the labeled antibodies. This allowed the antibodies to penetrate more deeply into the chromatin structure, and boosted the number of gold particles adhering to regions of interest. The signal was enhanced by adding a silver solution that precipitated (solidified) upon contact with the gold.
“We are interested in chromatin structure, so our targets are mostly chromatin-bound proteins,” Kireev said.
The researchers had inserted several copies of a bacterial DNA, called the Lac operator, into the chromosomes. A bacterial protein, the Lac repressor, recognizes and binds to the Lac operator in living cells.
The researchers combined a Lac repressor protein with another protein that fluoresces green under blue light. This engineered protein adhered to the chromosomes in regions containing the Lac operator sequences. Under blue light, these regions fluoresced. A gold-tagged antibody targeted against green fluorescent protein (GFP) was then microinjected into the nucleus of a living cell, which added a metallic signal that could be boosted with silver.
“All this combined gives us a much better signal, a much stronger signal, with the very best structural preservation,” Kireev said.
The fluorescing protein helped the researchers find the regions of interest in the cells. These areas were then “immunogold” labeled and targeted for electron microscopy.
In the resulting micrographs the researchers saw enhanced staining of the chromosomes.
“We can now apply this same live-cell labeling method to study at high resolution many different GFP-tagged proteins in the cell cytoplasm or nucleus,” said Andrew Belmont, a professor of cell and developmental biology and senior author of the paper.
“In trying to understand chromosomes, people have largely been limited to low resolution visualization of specific chromosomal proteins using light microscopy,” Belmost said. “This meant everyone has had to do a lot of guessing of how things are put together, leading in many cases to vague, cartoon models of what are likely to be complicated chromosomal structures carrying out DNA functions such as replication and transcription.”
“Now we hope we can simply look and see the real structure using the more than 10-fold higher resolution of electron microscopy,” Belmont said. “We are really excited to see what we will find using our new method”
Source : University of Illinois at Urbana-Champaign
 Print Article
Print Article Mail to a Friend
Mail to a Friend
